学位論文要旨
| No | 123003 | |
| 著者(漢字) | 臧,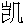  | |
| 著者(英字) | ||
| 著者(カナ) | ザン,カイサイ | |
| 標題(和) | 活性汚泥中に存在するエストロン分解細菌のマイクロオートラジオグラフィー蛍光in situハイブリダイゼーション法を用いた系統学的同定 | |
| 標題(洋) | Phylogenetic identification of estrone-degrading bacteria in activated sludge using microautoradiography-fluorescence in situ hybridization | |
| 報告番号 | 123003 | |
| 報告番号 | 甲23003 | |
| 学位授与日 | 2007.09.28 | |
| 学位種別 | 課程博士 | |
| 学位種類 | 博士(工学) | |
| 学位記番号 | 博工第6620号 | |
| 研究科 | 工学系研究科 | |
| 専攻 | 都市工学専攻 | |
| 論文審査委員 | ||
| 内容要旨 | Endocrine disrupting chemicals (EDCs) are described as the exogenous substances that alter function(s) of the endocrine system and consequently cause adverse health effects in an intact organism, or its progeny, or (sub)-populations (WHO/IPCS, 2002). In the last 20 years, a series of studies reported the reproductive changes in various species of fish and mollusks living downstream of sewage treatment plant (STP) outfalls (Howell et al., 1980, Bortone and Davis, 1994, Purdom et al. 1994, Tyler et al., 1998, Irwin et al., 2001 and Gagne et al., 2002). The causative compounds were suggested to be the EDCs in sewage effluents such as 4-tertiary isomers of nonylphenol and octylphenol, natural and synthetic estrogen including 17β-estradiol (E2), estrone (E1), estriol (E3) and 17α-ethinylestradiol (EE2). Although a variety of EDCs have been observed in sewage effluents, the further toxicity identifications or evaluations of EDCs in sewage effluents suggested that natural estrogens and occasionally the synthetic estrogen were the main contributors to the estrogenic potency of sewage effluents (Desbrow et al., 1998, Rodgers-Gray et al., 2000 and Aerni et al., 2004). The natural estrogens after discharged by households are transported to STPs, where ideally, they are completely degraded before discharged to water environments. However, the removal efficiency of natural estrogens in real STPs varies greatly (Fujii et al., 2002; Joss et al., 2004; Johnson et al., 2005 and Auriol et al., 2006). So far, the reasons causing insufficient estrogen-removal are still unclear. It is likely that longer HRT and SRT contribute to better removal (Saino et al., 2004 and Johnson et al., 2005). Such findings indicated the importance of biological activity to estrogen removal. To date, a number of bacterial strains have been isolated from activated sludge or soil, which can utilize E1, E2, E3 or even EE2 as carbon sources. Such isolates include Novosphingobium tardaugens sp. nov (Fujii et al., 2002), Rhodococcus zopfii (Yoshimoto et al., 2004), Rhodococcus equi (Yoshimoto et al., 2004), Alcaligenes xylosoxidans (Weber et al., 2005), Ralstonia sp (Weber et al., 2005) and Denitratisoma oestradiolicum (Fahrbach et al., 2006). In addition, Nitrosomonas europaeaーwell-known ammonia-oxidizing bacteria isolate was also proved capable of degrading E1 E2, E3 and EE2 to a certain degree by co-metabolic activity (Shi et al., 2004). It is still unclear, however, who are the key members responsible for estrogen degradation in a complex microbial community such as activated sludge. This problem can be partially addressed using microautoradiography-fluorescence in situ hybridization (MAR-FISH) analysis. MAR-FISH, as a powerful tool to directly link phylogenetic identity of a cell to its specific metabolic activity in a microbial community, has successfully analyzed the phylogeny and in situ physiology of microbial communities in many studies. So far, most MAR-FISH studies carried out are related to the nutrient uptake activities of microbial communities in different environments, and very limited work have been done to investigate the micropollutant-uptake activity by a microbial community in their habitat. The aim of this study was to explore the bacteria responsible for the in situ E1 degradation in activated sludge, using the improved MAR-FISH. In preliminary tests, slightly modified MAR-FISH technique with [3H]E1 and [3H]E1-3-sulfate conjugate ([3H]E1-S) as radioisotope tracers was firstly applied to E1- and/or E1-S-degrading pure cultures Rhodococcus sp. ED7 and Sphingomonas sp. ED8 to demonstrate the feasibility of MAR-FISH for in situ analysis of E1- and E1-S-degrading bacteria. Before the validation, a slight modification of the washing step after MAR incubation in MAR-FISH analytical procedure was made in order to remove the adsorbed radioactive substrates from biomass more efficiently. In the first stage of this study, the uptakes of [3H]E1 and [3H]E1-S by the major phylogenetic groups in activated sludge samples were investigated using MAR-FISH with broad group specific oligonucleotide probes. The activated sludge samples were taken from the anaerobic-anoxic-oxic process from two municipal wastewater treatment plants locating in Tokyo, Japan. Each sludge sample was incubated with 200 μg/L of tritium-labeled and unlabeled E1 mix. During E1 degradation process, the distribution patterns of tritium-assimilating cells among the major phylogenetic groups at different times were examined. In the early phase of E1 degradation, about 1-2% of total bacterial community in both activated sludge samples were detected as [3H]E1-assimilating cells. Around 60-80% of EUBmix-defined [3H]E1-assimilating cells were hybridized with probe Bet42a for β-proteobacteria and 40-20% hybridized with probe GAM42a for γ-proteobacteria. Under the microscope, the betaproteobacterial MAR positive (+) cells were mostly rods in chain, but sometimes rods. While the gammaproteobacterial MAR (+) cells were in spherical or short rod shapes. Alphaproteobacterial tritium-assimilating cells were only detected in the late phase of E1 degradation. By the time of 90% of added E1 degraded, no tritium-assimilating cell associated with Actinobacteria, Nitrospira, Planctomycetes and Cytophaga-flavobacter cluster was detected. In the end of E1 degradation, a significant potion of tritium-assimilating filamentous cells related to Chloroflexi was detected in both sludge samples, which was likely caused by the cross-feeding of radioactive metabolites or microbial products derived from tritium-labeled bacteria. Similarly, the investigation of E1-S degradation in activated sludge samples revealed that the betaproteobacterial MAR (+) cells with the same morphotypes accounted for the biggest portion in the total tritium-assimilating cell communities, while gammaproteobacterial MAR (+) cells ranked second. In summary, both beta- and gamma-proteobacterial MAR (+) cells contributed the majority of [3H]E1- or [3H]E1-S-assimilating cells in the studied activated sludge samples. Betaproteobacterial MAR (+) cells contributed most to the in situ E1 and E1-S degradations. In the second stage of this study, a set of hierarchic oligonucleotide probes targeting the subgroups of β-proteobacteria were applied in MAR-FISH, analyzing activated sludge M in order to target the most important in situ E1-degrading bacteria in more and more specific phylogenetic level. MAR-FISH analysis revealed that around 99% of betaproteobacterial MAR (+) cells were hybridized with probe BONE, which targets β1 subgroup of Proteobacteria. No MAR (+) cells was observed binding of probe BTWO which targets most members in order Rhodocyclales. The further screening for MAR (+) cells within β1 group revealed probe Cte, which mainly targets the members in family Comamonadaceae and Incertae sedis-group, targeted betaproteobacterial MAR (+) cells as efficiently as probe BONE. Subsequent screening for MAR (+) cells was carried out within family Comamonadaceae and Incertae sedis-group with more specific probes (or probe mix). The applied probes include probe ACI208 targeting genus Acidovorax; probe mix COM1424 and CteA targeting genus Comamonas; probe DEN220 targeting acetate-denitrifying bacteria affiliated with genera Acidovorax and Comamonas and some others in Comamonadaceae family; probe PSP-6 targeting genera Aquabacterium, Leptothrix, Rubrivivax and Ideonella in Incertae sedis-group, and probe PS-1 targeting four isolated Leptothrix spp. However, MAR-FISH analysis revealed that none of them could target the betaproteobacterial MAR (+) cells of interest. In order to design oligonucleotide probes to target the betaproteobacterial MAR (+) cells of interest in activated sludge samples, DNA extracted from the same activated sludge sample was amplified by PCR with β-proteobacteria-specified primer set, followed by cloning and sequencing. A 690bp-16S rRNA clone library was constructed which contained 175 cloned sequences. One hundred and forty-seven cloned sequences out of 175 were different from each other. Of 175 cloned sequences, 129 of them affiliated with β-subclass of Proteobacteria. The retrieved β-proteobacteria-associated sequences were diverse in phylogenetic identity. Within β-subclass of Proteobacteria, the highest number of cloned sequences affiliated with order Burkholderiales (71 clones) which was supposed to be targeted by probe BONE in hybridization. Order Rhodocyclales ranked second with 38 associated clones and followed by Nitrosomonadales, Hydrogenophilales, Methyloversatilis and Neisseriales. Most members in order Rhodocyclales, but not Zoogloea and Dechlorosoma-Azospira clusters were supposed to be targeted by probe BTWO. Within order Burkholderiales, 52 cloned sequences were grouped to family Comamonadaceae, and 12 cloned sequences to Incertae sedis-group. Besides, three cloned sequences were affiliated with family Alcaligenaceae and two cloned sequences affiliated with family Burkholderiaceae. Four oligonucleotide probes were designed according to the cloned sequences information, and used in fluorescence in situ hybridization to target most members in Incertae sedis-group (probe Inc1352); members in genera Sphaerotilus, Leptothrix, Ideonella and Schlegelella in Incertae sedis-group (probe Inc1197); and the cloned sequences affiliated with Sphaerotilus (probes Spha823 and Spha1037). Finally, MAR-FISH results with newly designed oligonucleotide probes revealed that the key in situ E1-degrading bacteria in the studied activated sludge samples were affiliated with the sheathed Sphaerotilus in Incertae sedis-group. Sphaerotilus related E1-degrading bacteria contributed 60 to 80 % of [3H]E1-assimilating cells during E1 degradations; and 78% of [3H]E1-S-assimilating cells during E1-S degradation in activated sludge samples. In summary, the in situ phylogenetic affiliation study of E1-degading bacteria in activated sludge revealed that betaproteobacterial E1-degrading bacteria contributed most to the in situ E1 and E-S degradations in the studied activated sludge samples and followed by gammaproteobacterial E1-degrading bacteria. MAR-FISH analysis combined with rRNA gene library survey successfully identified the most important E1-degrading bacteria as the sheathed Sphaerotilus with the newly designed probe Spha823. Sphaerotilus-related E1-degrading bacteria were phylogenetically different from all the previously reported E1-degrading bacterial isolates. They were proved as contributing most to the E1 and E1-S degradations in the studied activated sludge samples. The successful identification of the key in situ E1- and E1-S-degrading bacteria in activated sludge proved the feasibility of MAR-FISH technique as a discovery tool if coupled with 16S rRNA gene library survey to discover the bacteria with specific function. | |
| 審査要旨 | 本研究は、「Phylogenetic identification of estrone-degrading bacteria in activated sludge using microautoradiography-fluorescence in situ hybridization」と題して、8つの章から論文を構成している。 第1章では、研究の背景と目的、および論文の構成を述べている。 第2章では、環境中におけるエストロン(E1)やエストラジオールなどのエストロゲン物質に起因する環境問題や活性汚泥法におけるエストロゲン除去について整理している。また、過去に報告されている活性汚泥中や土壌中のE1分解細菌の分類整理をしている。 第3章では、都市下水処理場から採取した活性汚泥、化学分析手法や培養実験方法を説明している。また、細菌群の系統学的分類のための手法としてのマイクロオートラジオグラフィー蛍光in situ ハイブリダイゼーション(MAR-FISH)法の原理や、エストロンのような微量汚染物質の分解に関与する細菌の同定への適用性を説明している。 第4章では、E1分解細菌であるRhodococcus sp. ED7 と Sphingomonas sp. ED8を対象に、MAR-FISH法によりエストロンを代謝していることを検出できるかどうかの確認している。また、比較的疎水性の高いエストロンを対象としていることから、MAR-FISH法を適用する上で、細菌に付着した余剰な標識エストロンの洗浄方法を検討した結果、超音波処理を含めた改良洗浄方法を提案している。 第5章では、異なる活性汚泥を対象に、エストロンを添加する分解培養実験を実施してE1分解速度やその特性をまず把握している。そして、重水素で標識したエストロンを分解している活性汚泥に対し、α、β、γ-proteobacteria など8種類の細菌群に特異的なプローブを利用したMAR-FISH法を適用することにより、どの細菌群にエストロン分解に関与する細菌が存在するかの確認を行った。その結果、活性汚泥中の約1-2%の細菌群がエストロン分解に関与していること、添加直後(6時間培養)ではβ-及びγ-proteobacteriaが、その後(10時間培養)α-proteobacteriaやChloroflexi門に属する細菌が標識されていることが判明していることを報告している。特に、培養初期により多く標識が観察されたβ-proteobacteriaが、最も重要なE1分解細菌を含んでいること、次いでγ-proteobacteria、さらに重水素で標識した硫酸抱合体エストロンを投与した試験で検出されたα-proteobacteriaもE1分解細菌として重要な細菌を含んでいること考察している。 第6章では、E1分解細菌のうち、最も重要と想定されたβ-proteobacteriaに含まれるエストロン分解細菌の絞り込みを行うことを試みている。階層的に特異的なプローブを利用したMAR-FISH法を繰り返し行うことにより、エストロン分解に関与している細菌群を探索した結果を示している。まず、β-proteobacteria内を階層的にとらえる既存のプローブを用い、MAR陽性の細菌を包含する一群を、Burkholderiales目に含まれるComamonadaceae科、およびIncertae cedis群の一群であると絞り込んだ。次に、活性汚泥からβ-proteobacteriaのクローンライブラリを179クローン分作成し、上記グループに属するクローンを得た。そして、最終的にSphaerotilus 属近縁の9クローンを含むSHA1036とSHA822プローブを新たに設計し、そのプローブにより、活性汚泥中のβ-proteobacteria に属するMAR陽性の細菌のうち98%を実験的に検出できることを示している。したがって、このプローブが活性汚泥中のE1分解細菌を評価できる手法に発展できるものと結論付けている。また、これらの分解細菌の形態は、鎖状あるいは分散した状態で存在する桿菌であったことも観察している。 第7章では、6章で設計したSphaerotilus 属近縁 E1分解細菌を検出するためのオリゴヌクレオチドプローブを用い、重水素で標識したエストロンおよびその硫酸抱合体を添加した3種の活性汚泥を対象に、エストロン分解に寄与している細菌におけるSphaerotilus 近縁 E1分解細菌の割合を調べている。その結果、それぞれ60%、80%および78%を占めていることを実験的に示している。高濃度のエストロン添加条件であるものの、このSphaerotilus 近縁 E1分解細菌が活性汚泥プロセスにおいてエストロン分解に深く寄与していることを示唆する知見を得ている。 第8章では、上記の研究成果から導かれる結論と今後の課題や展望が述べられている。 以上の成果では、活性汚泥処理におけるエストロン分解に関わる微生物群集に関する知見を提供しただけでなく、エストロン分解に関与していると想定される微生物を検出できる配列特異的なオリゴヌクレオチドプローブを設計し、その有用性を示している。また、MAR-FISH法と16S rRNA遺伝子ライブラリー検索を組み合わせることで、階層的に特異的なFISHプローブを設計して、特定の物質を分解する細菌を絞り込む手順を提示するなど、複合微生物系の群集解析手法において非常に有用なデータや知見を提供しており、都市環境工学の学術の進展に大きく寄与するものである。 よって,本論文は博士(工学)の学位請求論文として合格と認められる。 | |
| UTokyo Repositoryリンク |